| The task of this project is to create the universal XML viewer component,
suitable for displaying different XML tree fragments in bioinformatical application. The
main class, representing such component, is Ulm.XML.GUI.XML_view. This component can also perform syntax analysis and display a structure of the given document. Despite this is mainly the java bean for programmers, it comes with the complete additional application that can view XML files. Hence the project is also useful for the end user. The
project was initiated by lack of the freely
available good XML viewer component to display data and results and do even simpliest analysis on the actual structure (not what is on DTD) of the existing XML document. Bioinformatical documents
also have many specifical data structures that can be displayed more clearly than just
representing XML tags. It is typical to have many nested tags, containing only one
other tag inside without additional information, for example:
<Auth-list>
<Auth-list_names>
<Auth-list_names_std>
<Author>
<Author_name>
<Person-id>
<Person-id_name>
<Name-std>
<Name-std_last>Xia</Name-std_last> ....
When you are simply showing such structure in a form of tree, no matter, collapsed or
uncollapsed, it is rather difficult to view. Our viewer detects such cases and displays
the simple nested tags in one line when possible. Some other
improvements also were suggested to improve readability.
This component has two additional putative fields. One of them allows to set the filter:
only the set of document fragments each having the indicated tag on the top is shown. The
other component is displaying the full path to the currently selected node. This
path is needed when programming XSLT and is shown correctly even if some root
nodes are hidden by the current filter. Finally, after setting the StructureVisible property, the structure tab appears. |
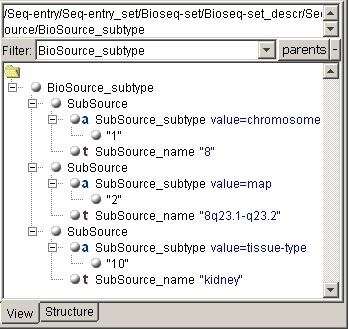
Structure tab:
 jar
executable for this test application. |

